Manual
We set the maximum sequence length limit for online submission to 1000 bases for H-type pseudoknots and to 400 bases if kissing hairpins are included. If you want to run DotKnot for longer sequences, please download the source code for installation on your local machine.
Input Format
Please provide a sequence ID for future reference. Paste your sequence in plain format into the appropriate field. Please ensure that only bases A,C,G,U,T (a,c,g,u,t) are used. If you would like to be notified when your computation is done, also give your email address (optional).
Example
Output Format
Approximate running times for DotKnot are in the order of seconds. For sequences close to 1000 bases, running time can be in the order of a couple of minutes. DotKnot will return the detected pseudoknots (if any) in the following format. Each pseudoknot is displayed in dot-bracket notation. Unpaired bases are indicated by dots. Base pairs are written as bracket pairs. The first stem of a pseudoknot is indicated by round brackets , i.e '(' and ')'. The second stem of a pseudoknot is indicated by square brackets, i.e. '[' and ']'. We also give the start and end positions of the pseudoknot with respect to the input sequence.
The user may wish to see a number of near-optimal local pseudoknots or the predicted global structure for the given sequence.
Example
HIV-1-RT
1 ... 35
UCAAGUAUUCCGAAGCUCAACGGGAAAAUGAGCUA
Position: 8 ... 34
UUCCGAAGCUCAACGGGAAAAUGAGCU
(((((.[[[[[[.)))))...]]]]]]
Estimated free energy: -15.1 kcal/mol
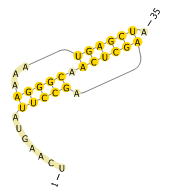
Visualization
For visualization of RNA secondary structures with pseudoknots, we recommend using
PseudoViewer. PseudoViewer supports
the dot-bracket notation as input format. For our example, the
2d-drawing looks like this: